How a novel machine learning system is bridging the gap between advanced models and expert insights to accelerate protein research
Proteins are essential molecules with many critical roles in the body. Determining a protein’s 3D structure helps scientists understand its functions within cells. This is crucial for advances in biology, medicine, and drug design.
Our paper published in Scientific Data introduces a novel AI-human hybrid workflow, developed by PBD and Europe PMC. The workflow identifies links between protein structure and function. The data, models and code are freely available.
Human expertise and AI approach
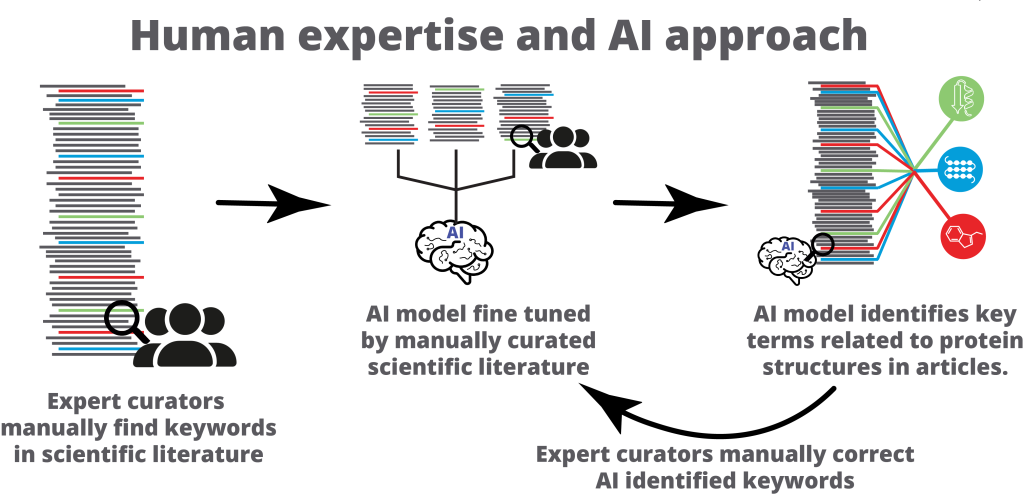
While human expertise is indispensable for its accuracy, it is often costly, time-consuming, and lacks scalability. On the other hand, AI offers speed and efficiency but requires expert input to train or fine-tune it to effectively perform complex tasks. This novel AI-human hybrid workflow combines the power of machine learning and expert curation to find links between protein structure and function.
The process starts with expert curators who manually review a set of scientific articles. They identify key terms related to protein structures. These key terms refer to the building blocks of proteins, known as amino acid residues. Residues’ specific locations and properties determine how the protein functions within the body.
The key terms identified from curation are then used to fine-tune a specialised AI model called PubMedBERT. The resulting model is highly specific, and key terms related to protein structures can be quickly found in articles. This helps scale the process of identifying the amino acid residue information and accelerates the discovery of connections between protein structures and their functions.
Accelerating discovery: faster and scalable annotations
The workflow developed by Europe PMC and PDBe bridges the gap between advanced machine learning and expert knowledge. The resulting model makes the identification of residues (the key terms) related to protein structure faster and easier to scale. The annotated terms it produces can be used to generate functional insights, support drug target validation, and propel the development of novel treatments.
“This work was a first start for PDBe in the field of NLP and the initial step for a more ambitious project downstream. The details given in the paper will hopefully allow anyone new to this field to follow through like in a recipe book.” Said Melanie Vollmar, PDBe Research Fellow (ARISE/Marie Curie).
Explore the article on Scientific Data to learn more about the AI-human approach and its potential impact on protein research.



