Europe PMC and Open Targets develop Lit-OTAR framework unearthing over 48 million unique associations that can be leveraged for drug discovery
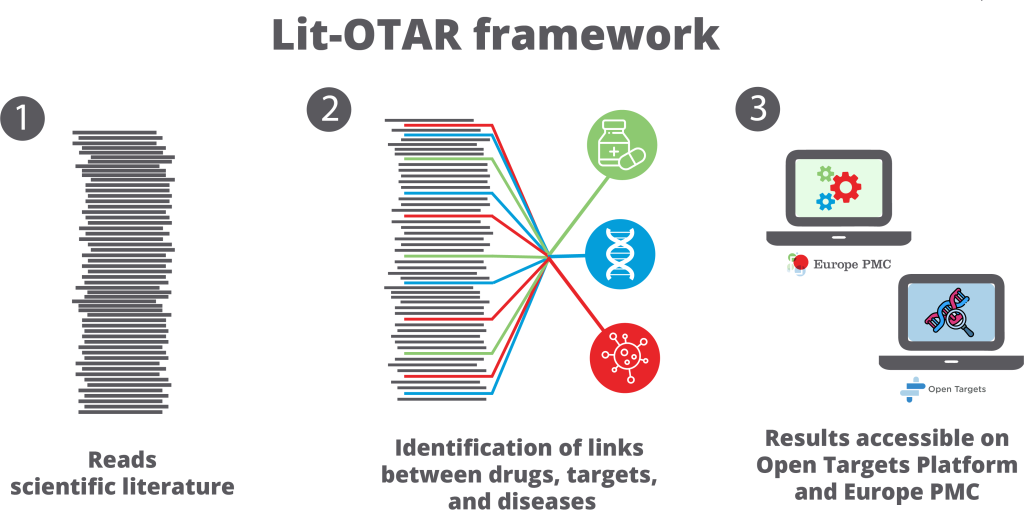
Identifying drug targets is a critical and intricate part of drug discovery. This requires scientists to look at many sources of evidence. They use them to find links between drugs, targets, and diseases. This helps them understand the underlying mechanisms of diseases and develop ideas for new treatments.
A recent preprint on bioRxiv introduces the Lit-OTAR framework, developed by Europe PMC and Open Targets. This framework uses deep learning to find connections that help identify drug targets, and the results are accessible on the Open Targets Platform and the Europe PMC website and Annotations API.
The Lit-OTAR framework
The Lit-OTAR framework scans scientific literature daily for gene – disease – drug associations. It extracts mentions of these entities from the text of research articles. The association is inferred from co-occurrence, meaning if a gene and a disease appear in the same sentence, they are likely connected. The algorithm also scores each article’s relevance to help prioritise potential targets.
Enhancing the drug discovery process
This approach provides comprehensive data for finding and validating drug targets. It speeds up the discovery of evidence for links between drugs, targets, and diseases, supporting the development of novel treatments.
“With the Lit-OTAR framework integrated into Europe PMC’s core infrastructure, our compute-efficient AI models continuously process new scientific articles, extracting gene, disease, and drug associations, enhancing research efficiency, supporting drug discovery, and advancing biocuration practices.” Said Santosh Tirunagari, Europe PMC Senior Machine Learning Developer.
Explore the full preprint on bioRxiv to learn more about the innovative Lit-OTAR framework and its potential impact on drug discovery.



